Convert experiment design into categorical colors
design2colors(
x,
group_colnames = NULL,
lightness_colnames = NULL,
class_colnames = NULL,
ignore_colnames = NULL,
preset = "dichromat2",
phase = 1,
rotate_phase = -1,
class_pad = 1,
end_hue_pad = 2,
hue_offset = 0,
desat = c(0, 0.4),
dex = c(2, 5),
Crange = c(70, 120),
Lrange = c(45, 90),
color_sub = NULL,
color_sub_max = NULL,
na_color = "grey75",
shuffle_colors = FALSE,
force_consistent_colors = TRUE,
plot_type = c("table", "list", "none"),
return_type = c("list", "df", "vector"),
verbose = FALSE,
debug = c("none", "cardinality"),
...
)Arguments
- x
data.framewith columns to be colorized,DataFramefrom BioconductorS4Vectorspackage,tbl_dffrom thetibblepackage, ormatrix. In all cases the data is coerced todata.framewithout changing colnames, and without imposing factor values per column, unless factors were already encoded.- group_colnames
characterorintgervector indicating whichcolnames(x)to use, in order, for group color assignment.- lightness_colnames
characterorintgervector indicating whichcolnames(x)to use, in order, for group lightness gradient.- class_colnames
characterorintgervector indicating higher-level grouping ofgroup_colnames- preset
characterstring passed tocolorjam::h2hwOptions(), which defines the hues around a color wheel, used when selecting categorical colors. Some shortcuts:"dichromat": (default) uses color-blindness friendly color wheel, minimizing effects of three types of color blindness mainly by removing large chunks of the green color space."ryb": red-yellow-blue color wheel, which emphasizes the yellow part of the wheel as a major color, as opposed to computer monitor default that represents the red-green-blue color components."ryb2": red-yellow-blue color wheel, version 2, adjusted to reduce effects of greenish-yellow for aesthetics."rgb": default red-green-blue color wheel used by computer monitors to mimic the components of human vision.
- phase, rotate_phase
integervalue,phaseis passed tocolorjam::rainbowJam()to define the light/dark pattern phasing, which has 6 positions, and negative values reverse the order. Categorical colors are assigned to the class/group combinations, after whichphase + rotate_phaseis used for categorical colors for any remaining values.- class_pad
integerzero or greater, indicating the number of empty hues to insert as a spacer between hues when the class changes in a sequence of class/group values. Higher values will ensure the hues in each class are more distinct from each other across class, and more similar to each other within class.- end_hue_pad
integerused to pad hues at the end of a color wheel sequence, typically useful to ensure the last color is not similar to the first color.- desat
numericvector extended to length=2, used to desaturate class/group colors, then remaining colors, in order. The intended effect is to have class/group colors visibly more colorful than remaining colors assigned to other factors.- dex
numericvector passed tojamba::color2gradient()to define the darkness expansion factor, where 1 applies a moderate effect, and higher values apply more dramatic light-to-dark effect. Whendexhas length=2, the second value is used only for columns where colors are assigned bycolnames(x)usingcolor_sub.- Crange, Lrange
numericranges passed tocolorjam::rainbowJam()to define slightly less saturated colors than the default rainbow palette.- color_sub
charactervector of R colors, wherenames(color_sub)assign each color to a character string. It is intended to allow specific color assignments upfront.colnames(x): whennames(color_sub)matches a column name inx, the color is assigned to that color using a color gradient across the unique character values in that column. Values are assigned in order of their appearance inxunless the column is afactor, in which case colors are assigned tolevels.
- color_sub_max
numericoptional value used to define a fixed upper limit to a color gradient when a color is applied to anumericcolumn.When one value is defined for
color_sub_maxit is used for allnumericcolumns uniformly.When multiple values are defined for
color_sub_max, thennames(color_sub_max)are used to associate to the appropriate column matching withcolnames(x).
- na_color
characterstring with R color, used to assign a specific color toNAvalues. (This assignment is not yet implemented.)- shuffle_colors
logicalindicating whether to shuffle categorical color assignments for values not already assigned by class/group/lightness, nor bycolor_sub. The effect is that colors are less likely to be similar for adjacent column values.- force_consistent_colors
logicalindicating whether to force color substitutions across multiple columns, when those columns share one or more of the same values. Note: This scenario is most likely to occur when usingcolor_subto assign colors to a specific column incolnames(x), and where that column may contain one or more values already assigned a color earlier in the process. For example: class/group/lightness defines colors for these columns, then all columns are checked for cardinality with these color assignments. Columns not appearing incolor_subwill be colorized when their cardinality is compatible with group colors, otherwise specific values may be assigned colors viacolor_sub, then all remaining values are assigned categorical colors. This process defines colors for each column. The last step reassigns colors consistently using the first value-color association that appears in the list, to make sure all assignments are consistent. This last step is subject of the argumentforce_consistent_colors. In either case, the outputcolor_subwill only have one color assigned per value.Default
TRUE: When colors are being assigned to column valuescolor_sub, if the value had been assigned a color in a previous column, for example bygroup_colnamescolor assignment, then the first assignment is applied to all subsequent assignments.Optional
FALSE: Color assignments are re-used where applicable, except when overridden bycolor_subfor a particular column. In that case, color assignments are maintained for each specific column.
- plot_type
characterstring indicating a type of plot for results:"table": plots a color table equal to the inputdata.framewhere background cells indicate color assignments."list": plots colors defined for each column inxusingjamba::showColors()"none": no plot is produced
- return_type
characterstring indicating the data format to return:"list": alistof colors, named bycolnames(x)."df": adata.framein order ofxwith colors assigned to each cell."vector": acharactervector of R colors, named by assigned factor level.
- verbose
logicalindicating whether to print verbose output.- debug
characterstring used to enable detailed debugging output.- ...
additional arguments are passed to downstream functions.
Value
output depends upon argument return_type:
"list": returns alistof colors defined bycolnames(x), suitable for use withComplexHeatmap::HeatmapAnnotation()for example."df": returnsdata.frameof colors with same dimensions as the inputx. Suitable for use withjamba::imageByColors()for example."vector": returnscharactervector of R colors, whose names represent values inx, where the values should be substituted with the color. Suitable for use withggplot2::color_manual(values=colors).In all cases, the
attributes()of the returned object also includes colors in the other two formats:"color_sub","color_df", and"color_list".
Details
The general goal is to assign categorical colors relevant to the experimental design of an analysis. The basic logic:
Assign categorical colors to experimental groups.
Shade these colors light-to-dark based upon secondary factors.
For step 1 above, optionally assign similar color hues by class.
When there are multiple factors in a design, the general guidance:
Define
group_colnamesusing the first two factors in the design.Define
class_colnamesusing one of these two factors. Values ingroup_colnameswill be assigned rainbow categorical colors, with extra spacing between each class. Values in one class will be assigned similar color hues, for example one class may be red/orange, another class may be blue/purple.Optionally choose another factor to use as
lightness_colnames. When there are multiple unique values per group, they will be shaded from light to dark within the group color hue.
It is sometimes helpful to create a column for class_colnames,
for example when a series of treatments can be categorized
by the type of treatment (agonists, antagonists, inhibitors,
controls).
Franky, we tend to try a few combinations until the output seems
intuitive. Then we assign specific values from other columns
using color_sub. Typically for numeric columns we assign
a color to the colname, and for categorical colors we assign
colors to values in the column.
Version 0.0.69.900 and higher: When the cardinality of group/class values is not 1-to-many, either the group/class assignments are switched in order to create 1-to-many cardinality, or a combination of the two vectors is used to create the appropriate cardinality.
When no group_colnames or class_colnames are defined
By default, the unique rownames are used as if they were groups,
then colors are assigned using the same logic as usual. Any
other column whose values are 1-to-1 match with rownames will
inherit the same colors, otherwise character and factor
columns will be assigned categorical colors, and numeric
columns will be assigned a color gradient.
Categorical colors
At its simplest a set of groups can be assigned categorical colors.
colors should be visibly distinct from one another
colors should generally be distinct across forms of color-blindness
colors should be consistent across plots, figures, tables
Finally, colors may be pre-defined using a named vector of colors. These colors will be propagated to other entries in the table.
Light-to-dark gradient
The light-to-dark gradient is intended for ordered sub-divisions, for example:
across time points in a time series
across treatment doses in an ordered series
across ordered measurements first-to-last
Group class
The group classification is intended to assign color hues for similar groups:
antagonists, agonists, untreated
treated, untreated
wildtype, mutant form 1, mutant form 2, etc.
For example, antagonists may be assigned colors blue-to-purple; agonists may be assigned colors red-to-orange; with a pronounced color hue "gap" between antagonists and agonists.
Additional categorical color assignment
Finally, other annotations associated with samples are assigned categorical colors, visibly distinct from other color assignments.
For entries associated with only one design color, for example "Sample_ID", "Sample Name", "Lane Number", or "Well Number", they inherit the design color.
For entries associated with more than one design color, for example "Batch", "Date", or perhaps "Dose", they will be assigned a unique color.
additional annotations unique to design colors inherit the design colors
additional categorical colors should not duplicate existing colors
Future ideas
Assign "additional factors" to colors based upon
classCurrently "additional factors" are only tested by class_group and class_group_lightness.
It could be useful to test versus
classalone (if supplied)Goal would be to assign color hue using the mean color hue in the class.
Otherwise the class may be assigned a color inconsistent with the range of color hues.
Handle numeric columns by applying color gradient
A truly numeric column (not just integer index values) could use
circlize::colorRamp2()to apply color gradient
See also
Other jam color functions:
assign_numeric_colors(),
df_to_numcolors(),
mean_hue(),
print_color_list(),
quick_complement_color()
Examples
df <- data.frame(
genotype=rep(c("WT", "GeneAKO", "GeneBKO"), c(4, 8, 8)),
treatment=rep(rep(c("control", "treated"), each=2), 5),
class=rep(c("WT", "KO"), c(4, 16)),
time=c(rep("early", 4),
rep(rep(c("early", "late"), each=4), 2)))
df$sample_group <- jamba::pasteByRow(df[,c("genotype", "treatment", "time")])
df$sample_name <- jamba::makeNames(df$sample_group);
df$age <- sample(40:80, size=nrow(df));
df
#> genotype treatment class time sample_group
#> 1 WT control WT early WT_control_early
#> 2 WT control WT early WT_control_early
#> 3 WT treated WT early WT_treated_early
#> 4 WT treated WT early WT_treated_early
#> 5 GeneAKO control KO early GeneAKO_control_early
#> 6 GeneAKO control KO early GeneAKO_control_early
#> 7 GeneAKO treated KO early GeneAKO_treated_early
#> 8 GeneAKO treated KO early GeneAKO_treated_early
#> 9 GeneAKO control KO late GeneAKO_control_late
#> 10 GeneAKO control KO late GeneAKO_control_late
#> 11 GeneAKO treated KO late GeneAKO_treated_late
#> 12 GeneAKO treated KO late GeneAKO_treated_late
#> 13 GeneBKO control KO early GeneBKO_control_early
#> 14 GeneBKO control KO early GeneBKO_control_early
#> 15 GeneBKO treated KO early GeneBKO_treated_early
#> 16 GeneBKO treated KO early GeneBKO_treated_early
#> 17 GeneBKO control KO late GeneBKO_control_late
#> 18 GeneBKO control KO late GeneBKO_control_late
#> 19 GeneBKO treated KO late GeneBKO_treated_late
#> 20 GeneBKO treated KO late GeneBKO_treated_late
#> sample_name age
#> 1 WT_control_early_v1 76
#> 2 WT_control_early_v2 53
#> 3 WT_treated_early_v1 64
#> 4 WT_treated_early_v2 65
#> 5 GeneAKO_control_early_v1 66
#> 6 GeneAKO_control_early_v2 44
#> 7 GeneAKO_treated_early_v1 80
#> 8 GeneAKO_treated_early_v2 67
#> 9 GeneAKO_control_late_v1 48
#> 10 GeneAKO_control_late_v2 68
#> 11 GeneAKO_treated_late_v1 42
#> 12 GeneAKO_treated_late_v2 47
#> 13 GeneBKO_control_early_v1 77
#> 14 GeneBKO_control_early_v2 46
#> 15 GeneBKO_treated_early_v1 49
#> 16 GeneBKO_treated_early_v2 72
#> 17 GeneBKO_control_late_v1 58
#> 18 GeneBKO_control_late_v2 43
#> 19 GeneBKO_treated_late_v1 79
#> 20 GeneBKO_treated_late_v2 56
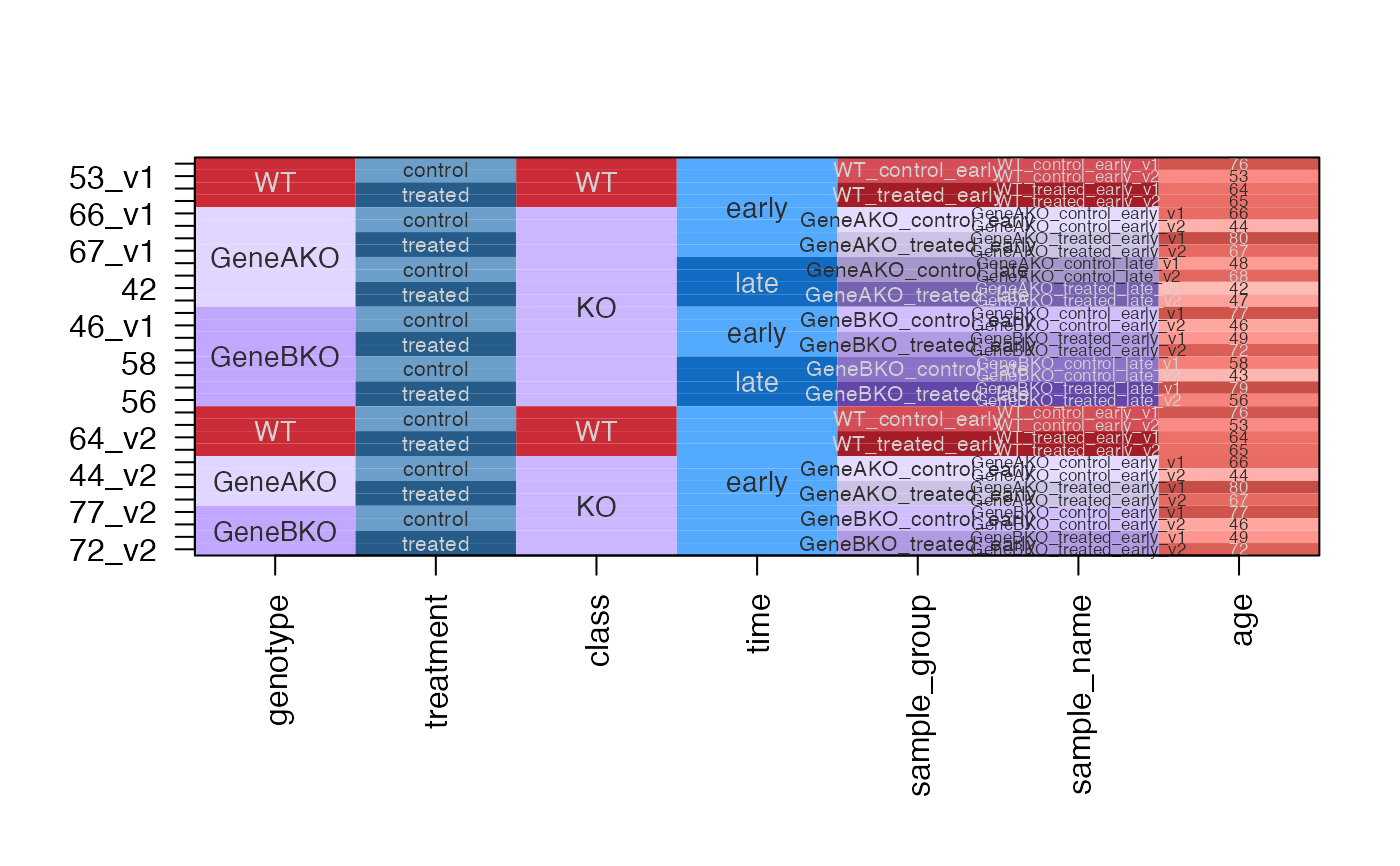
dfc <- design2colors(df,
group_colnames="genotype",
lightness_colnames="treatment",
class_colnames="class",
color_sub=c(age="dodgerblue"))
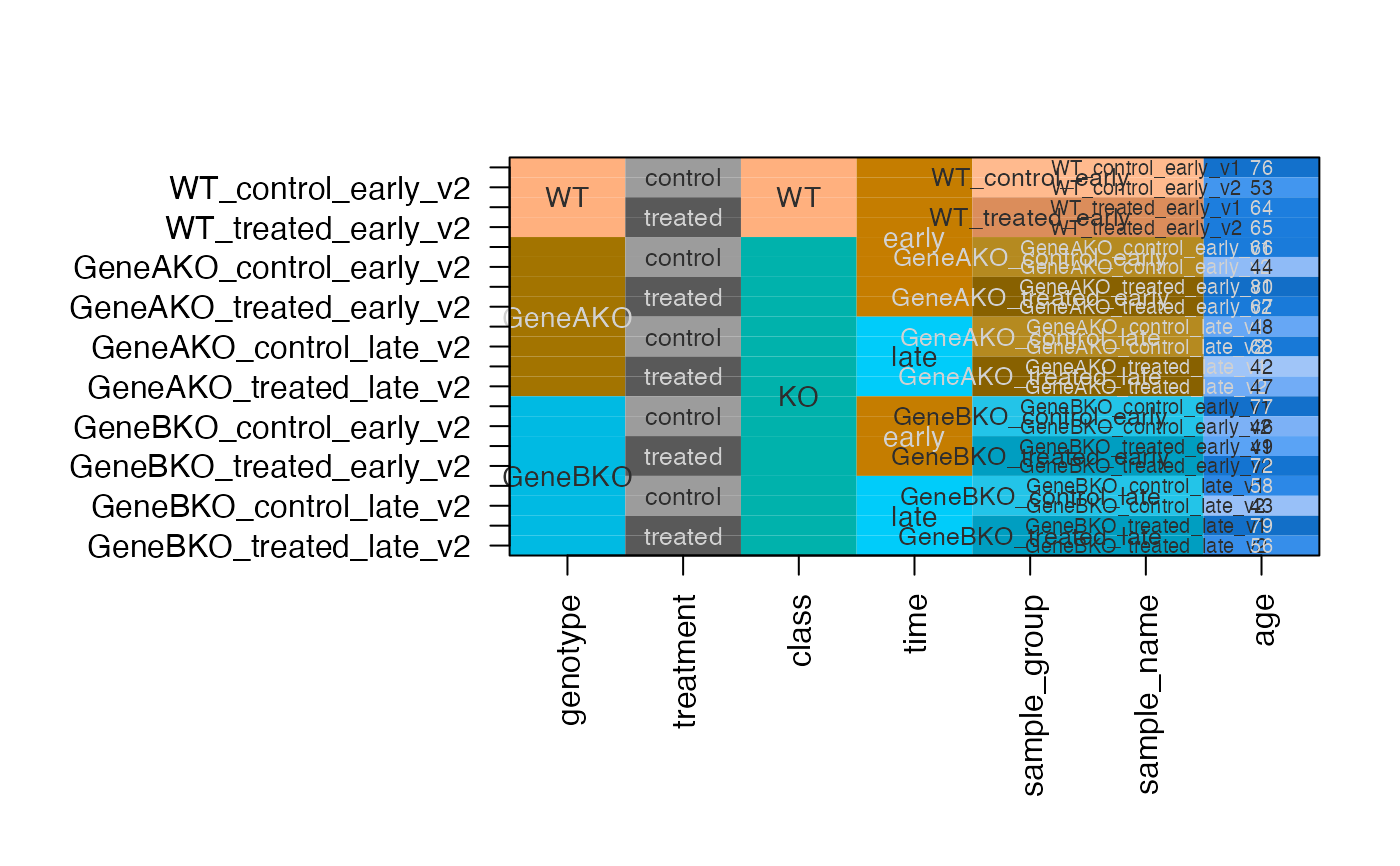 # same as above except assign colors to columns and some values
dfc <- design2colors(df,
group_colnames="genotype",
lightness_colnames="treatment",
class_colnames="class",
preset="dichromat",
color_sub=c(KO="firebrick3",
treatment="navy",
class="cyan",
time="dodgerblue"))
# same as above except assign colors to columns and some values
dfc <- design2colors(df,
group_colnames="genotype",
lightness_colnames="treatment",
class_colnames="class",
preset="dichromat",
color_sub=c(KO="firebrick3",
treatment="navy",
class="cyan",
time="dodgerblue"))
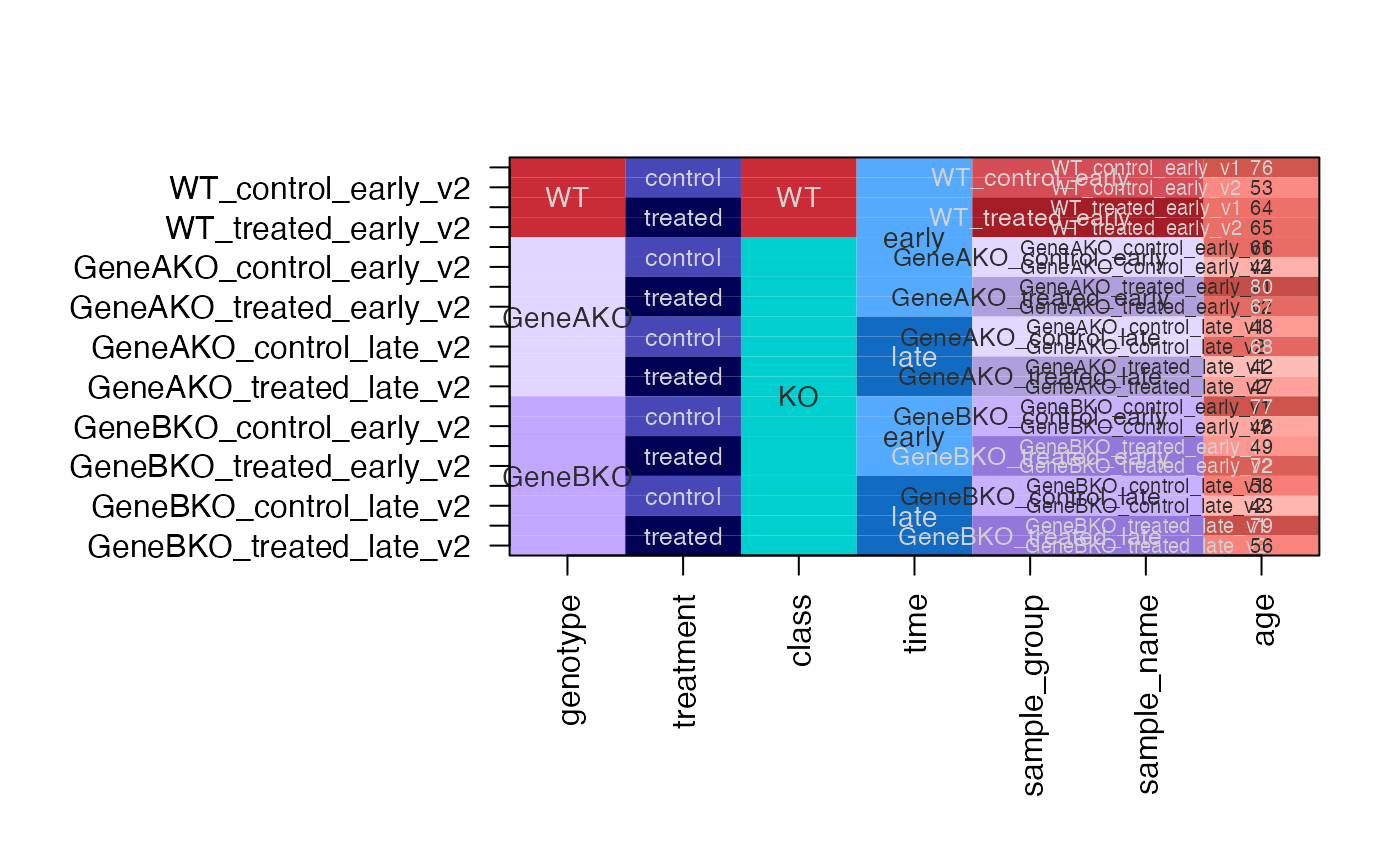 # same as above except assign specific group colors
dfc <- design2colors(df,
group_colnames="genotype",
lightness_colnames="treatment",
class_colnames="class",
preset="dichromat",
color_sub=c(
WT="gold",
KO="purple3",
GeneAKO="firebrick3",
GeneBKO="dodgerblue",
treatment="navy",
time="darkorchid4"))
# same as above except assign specific group colors
dfc <- design2colors(df,
group_colnames="genotype",
lightness_colnames="treatment",
class_colnames="class",
preset="dichromat",
color_sub=c(
WT="gold",
KO="purple3",
GeneAKO="firebrick3",
GeneBKO="dodgerblue",
treatment="navy",
time="darkorchid4"))
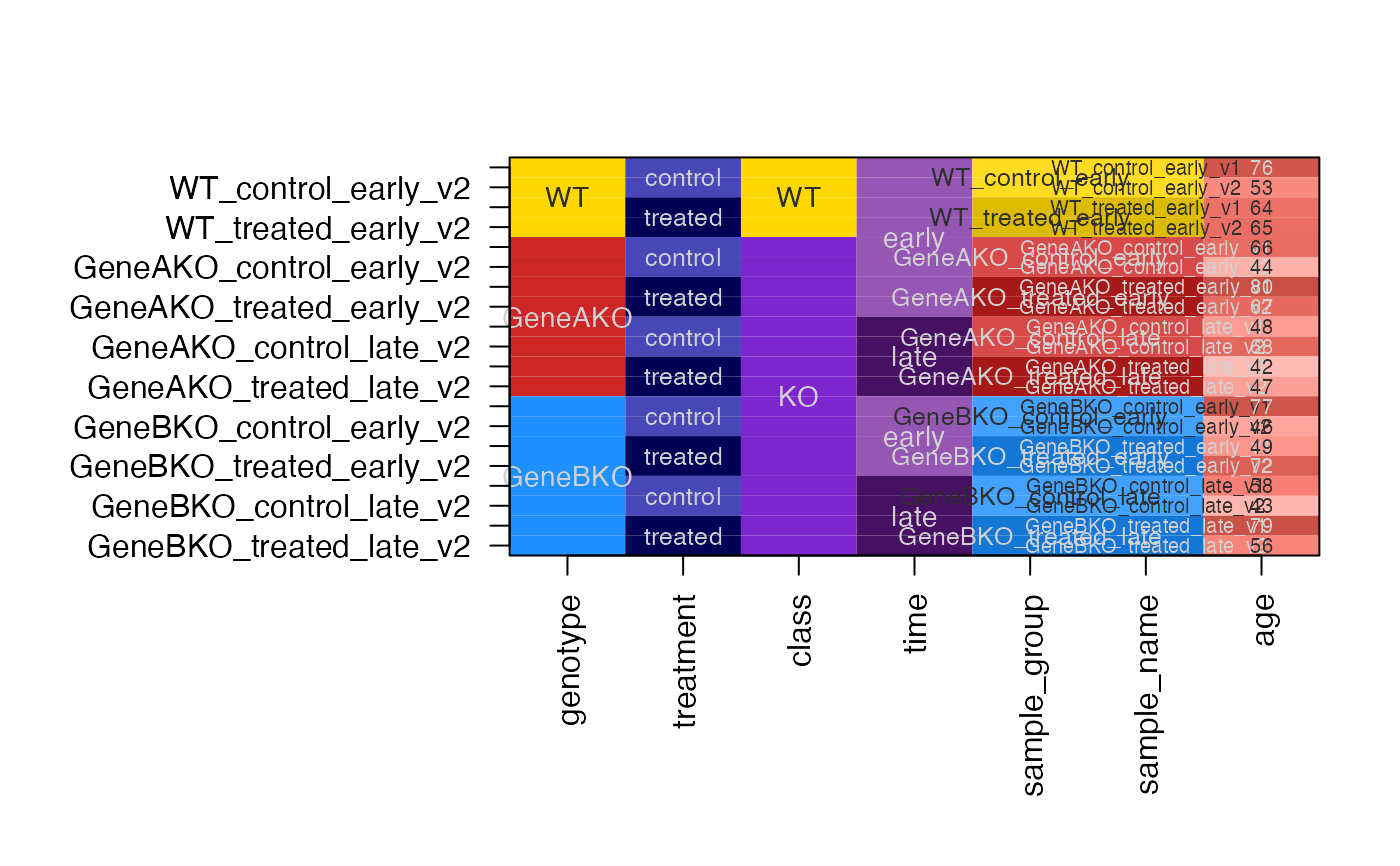 dfc2 <- design2colors(df,
group_colnames="genotype",
lightness_colnames=c("time", "treatment"),
class_colnames="class",
preset="dichromat")
dfc2 <- design2colors(df,
group_colnames="genotype",
lightness_colnames=c("time", "treatment"),
class_colnames="class",
preset="dichromat")
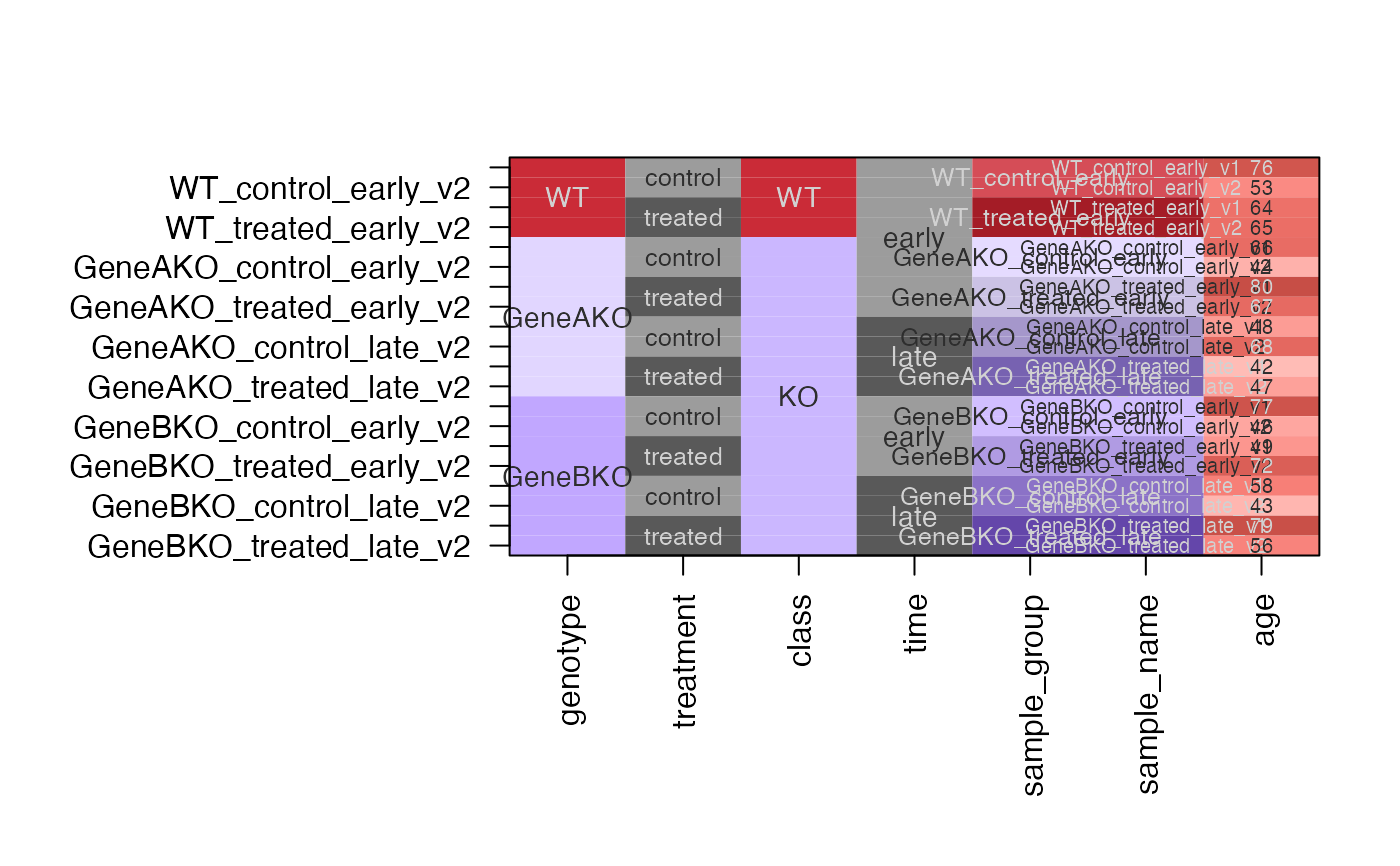 dfc3 <- design2colors(df,
group_colnames=c("genotype"),
lightness_colnames=c("time", "treatment"),
class_colnames="genotype",
rotate_phase=TRUE,
preset="dichromat")
dfc3 <- design2colors(df,
group_colnames=c("genotype"),
lightness_colnames=c("time", "treatment"),
class_colnames="genotype",
rotate_phase=TRUE,
preset="dichromat")
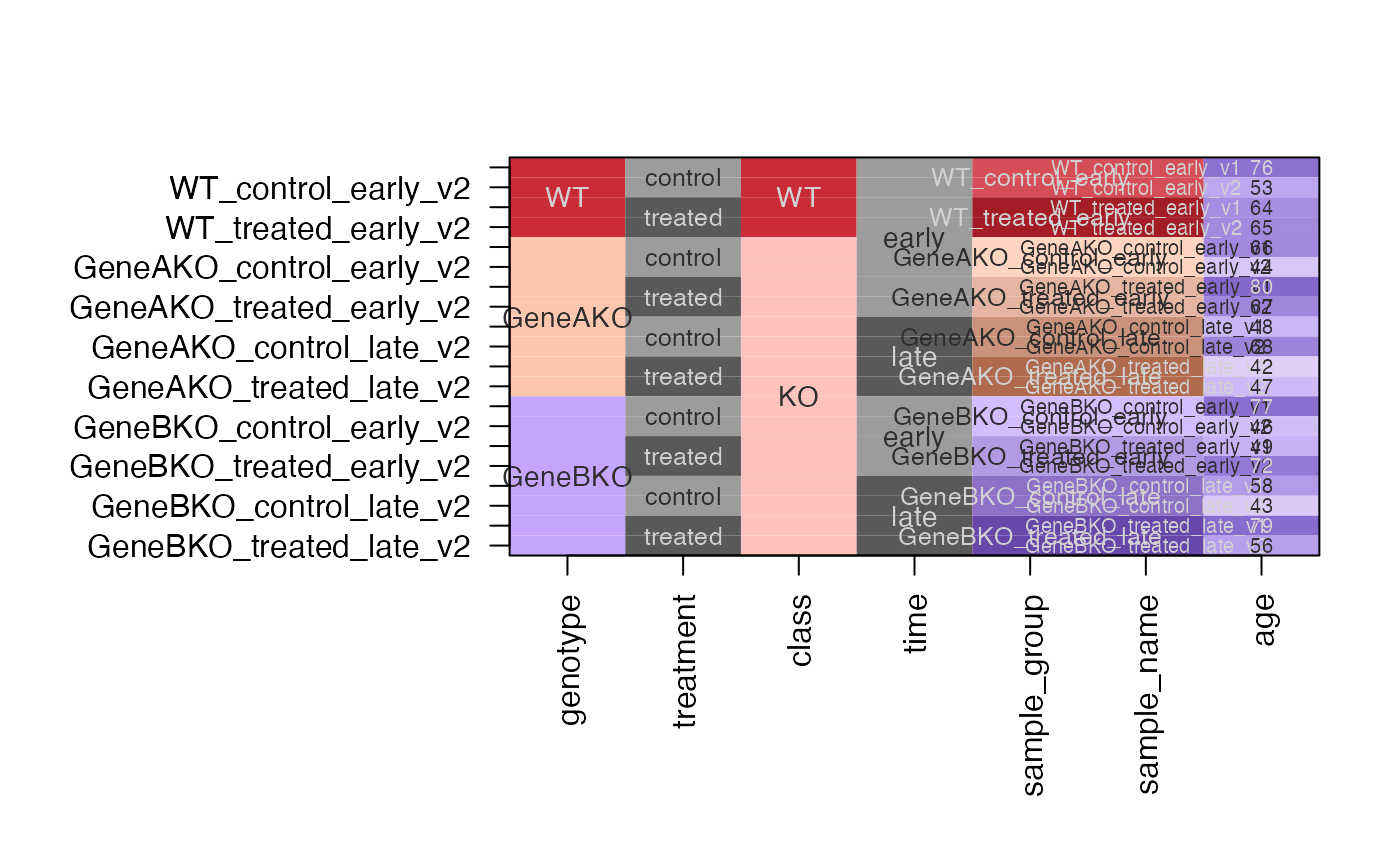 df1 <- df;
df2 <- subset(df, time %in% "early");
df12 <- rbind(df1, df2);
dfc12 <- design2colors(df12,
group_colnames="genotype",
lightness_colnames=c("time", "treatment"),
class_colnames="class",
preset="dichromat",
color_sub=c(
treatment="steelblue",
time="dodgerblue"
))
df1 <- df;
df2 <- subset(df, time %in% "early");
df12 <- rbind(df1, df2);
dfc12 <- design2colors(df12,
group_colnames="genotype",
lightness_colnames=c("time", "treatment"),
class_colnames="class",
preset="dichromat",
color_sub=c(
treatment="steelblue",
time="dodgerblue"
))