Relayout each nodegroup in a bipartite (Cnet) graph, experimental
Source:R/jamgraph-relayout-nodegroups.R
relayout_nodegroups.RdRelayout each nodegroup in a bipartite (Cnet) graph, experimental
Usage
relayout_nodegroups(
cnet,
nodegroups = NULL,
repulse = 3.5,
fix_set_nodes = TRUE,
spread_labels = TRUE,
add_edges = TRUE,
edge_factor = 2,
do_final_relayout = NULL,
final_repulse = 3.5,
apply_by_size = TRUE,
byCols = c("-num_terms", "-num_nodes", "nodegroup"),
verbose = FALSE,
...
)Arguments
- cnet
igraphobject with node layout already defined- nodegroups
listof node names, orcommunitiesobject, passed tocommunities2nodegroups(). When NULL, it checks for supporting data in this order:If graph attribute 'mark.groups' is defined, it is used.
If vertex attribute 'nodeType' exists, it calls
get_cnet_nodeset().Finally, it calls
igraph::cluster_optimal(), hoping this method will be appropriate for the graph size.
- repulse
numericdefault 3.5, passed torelayout_with_qfr()- fix_set_nodes
logicaldefault TRUE, whether to fix all nodes with nodeType=='Set' to prevent them from moving.- spread_labels
logicaldefault TRUE, whether to applyspread_igraph_labels()after the relayout iterations are complete.- add_edges
logicaldefault TRUE, whether to add edges within nodes of each nodegroup. The same is accomplished by settingedge_factor=0.- edge_factor
numericdefault 2, used as the numerator in new edge weights with equationedge_factor/sqrt(n)where 'n' is the number of nodes in the nodegroup.Set
edge_factor=0oradd_edges=FALSEto skip this step.
- do_final_relayout
logicaldefault NULL, whether to apply one morerelayout_with_qfr()after each nodegroup is adjusted. It usesfinal_repulse.- final_repulse
numericused whendo_final_relayoutis TRUE, used as the 'repulse' argument inrelayout_with_qfr().- apply_by_size
logicaldefault TRUE, whether to apply the relayout to nodegroups ordered by size, usingbyColsto sort.The default applies layout such that nodegroups with the most terms in
names(nodegroups)are applied first, then largest to smallest nodegroups.For Cnet plots, Gene nodes in nodegroups connected to the most Set nodes are adjusted first, then largest nodegroups, then sorted by nodegroup name.
It is unclear if the order is useful, future iterations of this approach may "move" other nodegroups aside first, then re-introduce each nodegroup into the layout one by one. Otherwise nodes could become "tangled" in the center, with no ideal method to optimize separation by nodegroup.
- byCols
charactervector used whenapply_by_sizeis TRUE.'-num_terms': reverse-order by the number of terms in each nodegroup name, assuming comma-delimited terms.
'-num_nodes': reverse-order by the number of nodes in each nodegroup.
'nodegroup': alphnumeric sort of the
names(nodegroups).
- verbose
logicalwhether to print verbose output.- ...
additional arguments are passed to
communities2nodegroups()for argument 'sep', or tospread_igraph_labels()for arguments regarding node ordering, etc.
Details
This function iteratively re-applies a layout function
to each nodegroup in a graph, constraining the position of
all other nodes for each iteration.
Currently the layout uses relayout_with_qfr(), in future it
may use any layout function.
The purpose is to "encourage" nodes in a nodegroup to become bundled together.
Strategy:
Each nodegroup is isolated, and
relayout_with_qfr()is called on nodes in each nodegroup, while constraining all other nodes so they cannot move.In theory, using the same
repulse, the nodes would not move at all. Changing the repulse force could encourage nodes to stay together.By default
add_edges=TRUEwhich adds phantom edges to connect all nodes in a node group.The edge weight is scaled down by the number of nodes.
These phantom edges are intended to help 'encourage' the nodegroup nodes to group together.
Otherwise, most layout algorithms are only focused on specific edge forces, and not secondary forces which are common in Cnet plots.
For example, nodes in a nodegroup all share the same network connections, however they are not otherwise attracted to each other in a network layout. In absence of any repulsive force, they would all be co-located. But with some repulsive force, they are repelled from each other, and sometimes end up radially positioned around the plot, and not grouped together.
The phantom edges add some minimal force for nodes to be grouped closer together, and are removed once the layout is complete.
As a final polishing step,
do_final_relayout=TRUEenables a final round of global node layout, withfinal_repulse.We observed that sometimes the nodegroups are too clumped, in a big circular "ball" due to the phantom edge process above. The final relayout is helpful to allow nodes to space out somewhat.
It may be helpful to pass
niterto control the number of layout iterations in this final step. The default is 500.
See also
Other jam cnet utilities:
adjust_cnet_nodeset(),
adjust_cnet_set_relayout_gene(),
apply_nodeset_spacing(),
bulk_cnet_adjustments(),
get_cnet_nodeset(),
get_cnet_nodeset_vector(),
launch_shinycat(),
make_cnet_test()
Examples
cnet <- make_cnet_test();
ns <- get_cnet_nodeset(cnet)
# mark.groups: highlight just one nodegroup
# nodegroups: enables the edge_bundling to work properly
jam_igraph(cnet, mark.groups=ns["SetA,SetB"], nodegroups=ns,
main="One nodegroup is split")
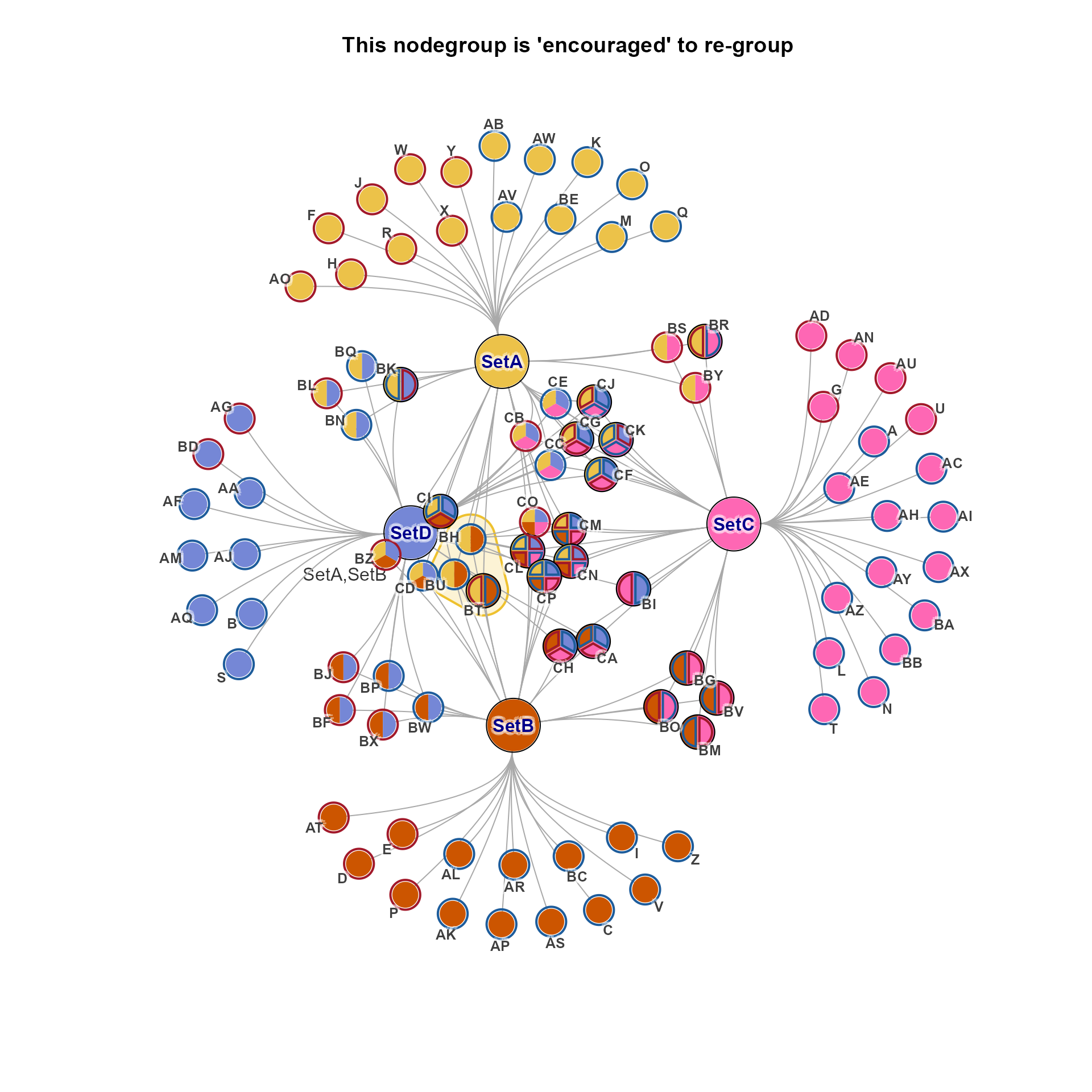
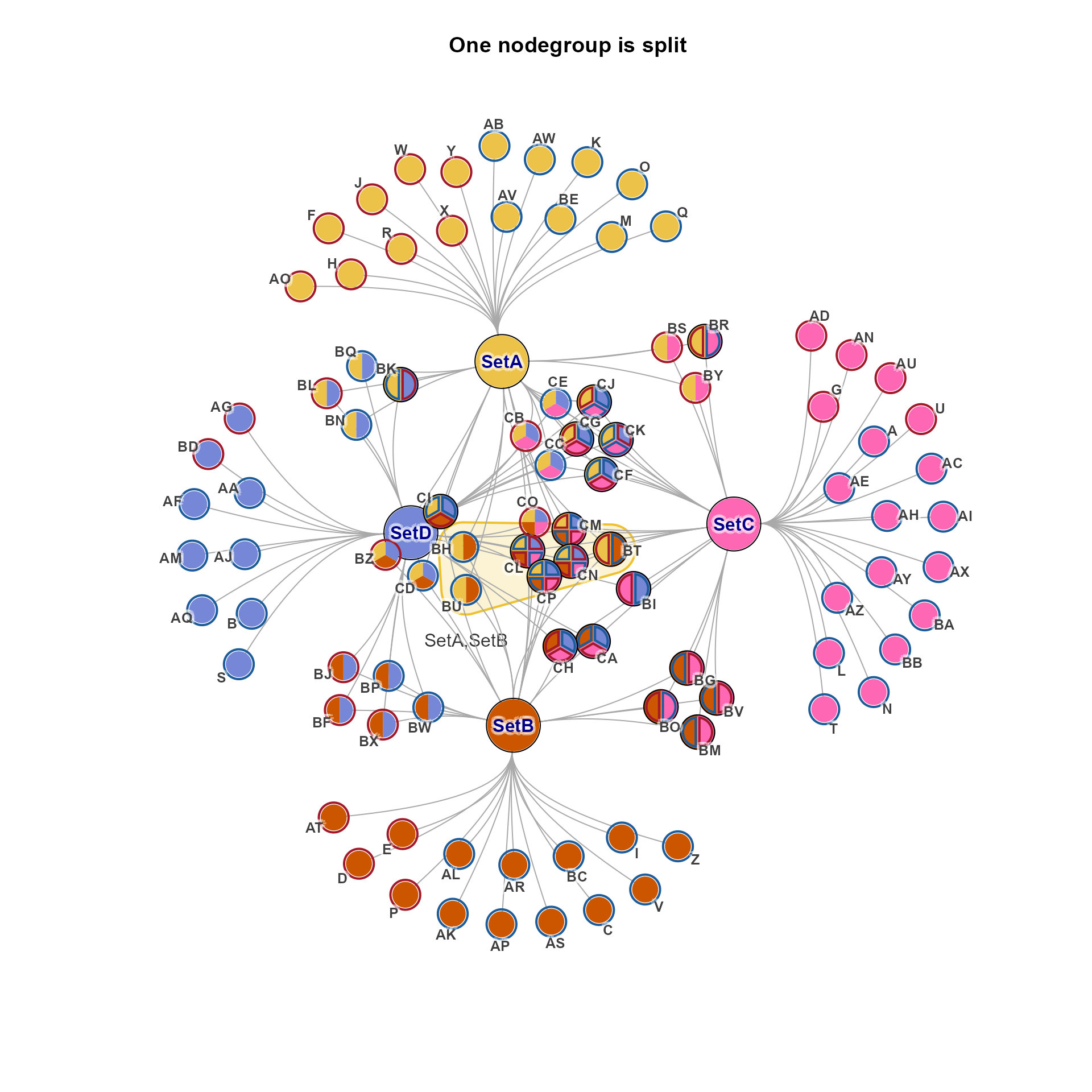 cnet2 <- relayout_nodegroups(cnet, nodegroups=ns["SetA,SetB"], do_final_layout=TRUE, verbose=TRUE)
#> ## (12:30:14) 16Dec2025: relayout_nodegroups(): Using nodegroups as supplied.
#> ## (12:30:14) 16Dec2025: relayout_nodegroups(): Applying to nodegroup 'SetA,SetB' with members: BH,BT,BU
jam_igraph(cnet2, mark.groups=ns["SetA,SetB"], nodegroups=ns,
main="This nodegroup is 'encouraged' to re-group")
cnet2 <- relayout_nodegroups(cnet, nodegroups=ns["SetA,SetB"], do_final_layout=TRUE, verbose=TRUE)
#> ## (12:30:14) 16Dec2025: relayout_nodegroups(): Using nodegroups as supplied.
#> ## (12:30:14) 16Dec2025: relayout_nodegroups(): Applying to nodegroup 'SetA,SetB' with members: BH,BT,BU
jam_igraph(cnet2, mark.groups=ns["SetA,SetB"], nodegroups=ns,
main="This nodegroup is 'encouraged' to re-group")
 # by default, it is applied to all nodes
cnet3 <- relayout_nodegroups(cnet)
jam_igraph(cnet3, mark.groups=ns, nodegroups=ns,
main="Re-layout across all nodegroups")
# by default, it is applied to all nodes
cnet3 <- relayout_nodegroups(cnet)
jam_igraph(cnet3, mark.groups=ns, nodegroups=ns,
main="Re-layout across all nodegroups")
