Convert multiEnrichMap mem output to EnrichmentMap emap
Usage
mem2emap(
mem,
overlap = 0.2,
p_cutoff = NULL,
min_count = NULL,
colorV = NULL,
cluster_function = igraph::cluster_walktrap,
cluster_list = NULL,
num_keep_terms = 3,
keep_terms_sep = ",\n",
repulse = 3.3,
remove_singlets = FALSE,
color_by_nodes = FALSE,
size_by_genes = TRUE,
median_size = 5,
apply_direction = TRUE,
direction_max = 2,
direction_floor = 0.5,
seed = 123,
spread_labels = TRUE,
label_min_dist = 2.5,
y_bias = 10,
vertex.label.font = 2,
use_shadowText = TRUE,
do_express = FALSE,
do_plot = FALSE,
verbose = FALSE,
...
)Arguments
- mem
Memor legacylistmem output frommultiEnrichMap()- overlap
numeric, default 0.2, value between 0 and 1 with Jaccard overlap coefficient required between any two pathways in order to create a network edge connecting these two pathways.Jaccard overlap coefficient is reciprocal of Jaccard distance, using
(1 - Jaccard_distance).overlap=0.2is default, which specifies roughly 20% overlap in genes shared between two pathway nodes.Note these genes must be involved in enrichment, and therefore does not use all possible genes annotated to a pathway. Therefore connections are only created with enriched genes are shared between pathways.
- p_cutoff
numericthreshold used for significant enrichment P-value. The default NULL uses the value in thememobject.- min_count
integerthreshold for minimum genes involved in enrichment in order for a pathway to be considered significant during this analysis. When NULL it uses the threshold inmem.- colorV
charactervector of R colors used for each enrichment. Default NULL uses colors defined inmem.- cluster_function
function, defaultigraph::cluster_walktrap, used to clusterigraphnodes in the resulting network graph. It is used to enhance the visual summary.This function is not used when
cluster_listis supplied.
- cluster_list
listdefault NULL, optional list of pathway clusters, containingcharactervectors ofigraphnode/vertex names. It will be used instead ofcluster_functionwhen supplied.- num_keep_terms
integer, default 3, number of text terms to keep for each pathway cluster, used together withcluster_function. Common words are removed, remaining terms are sorted by decreasing occurrence, then used to summarize each cluster.- keep_terms_sep
characterstring, default is comma-newline, used to separate terms, used together withnum_keep_terms.- repulse
numericvalue passed tolayout_with_qfr(), default 3.3.Use repulse 'FALSE', 'NULL', or '0' will skip the layout, which is used by
mem_find_overlap()to iterate numerous overlaps without spending time on layout each iteration.
- remove_singlets
logical, default TRUE, whether to remove pathway singlets which have no connections to any other pathways.Using TRUE will help simplify busy figures, at the expense of being less complete.
- color_by_nodes
logical, default TRUE, whether to colorize pathway clusters using node colors in each cluster. Note that a mix of colors often turns brown, so this feature has unpredictable benefit.- apply_direction
logicaldefault TRUE, whether to apply direction to the node border color, when supplied. Used withdirection_maxanddirection_floor.- direction_max
numeric, default 2, indicating the directional score at which the maximum color is applied. Typically when using some derivative of z-score to represent directionality, values at or above 2 are effectively "maximum".- direction_floor
numericdefault 0.5, the minimum directional score for any color to be applied, below which values are considered "non-directional". Typically when using some form of z-score, a value at or below 0.5 may be considered "not directional."- seed
numericpassed toset.seed()via thelayout_with_qfr()layout algorithm, default 123.- do_express
logicaldefault FALSE, when TRUE it skips a number of aesthetic steps. This option is intended mainly formem_find_overlap()to perform more rapid iterative evaluation of overlap thresholds.- do_plot
logical, default FALSE, whether to render the resulting plot usingjam_igraph().- verbose
logicalwhether to print verbose output.- ...
additional arguments are passed to
jam_igraph()to customize the network plot, used whendo_plot=TRUE.
Details
This function is currently In development.
This function is intended to convert mem output from
multiEnrichMap() into an EnrichmentMap igraph format
which represents the statistical enrichment support from each
pathway enrichment.
This function can apply P-value thresholds using the input mem,
or using a custom value.
A node clustering function is applied by default, which may help define suitable subgroups of nodes. When defined, the clusters are used to define nodegroups for edge bundling.
See also
Other jam Mem utilities:
Mem-class,
MemPlotFolio-class,
fixSetLabels(),
mem2cnet()
Other jam igraph functions:
communities2nodegroups(),
drawEllipse(),
edge_bundle_bipartite(),
edge_bundle_nodegroups(),
fixSetLabels(),
flip_edges(),
get_bipartite_nodeset(),
highlight_edges_by_node(),
igraph2pieGraph(),
label_communities(),
mem2cnet(),
nodegroups2communities(),
rectifyPiegraph(),
removeIgraphBlanks(),
subsetCnetIgraph(),
subset_igraph_components(),
sync_igraph_communities()
Examples
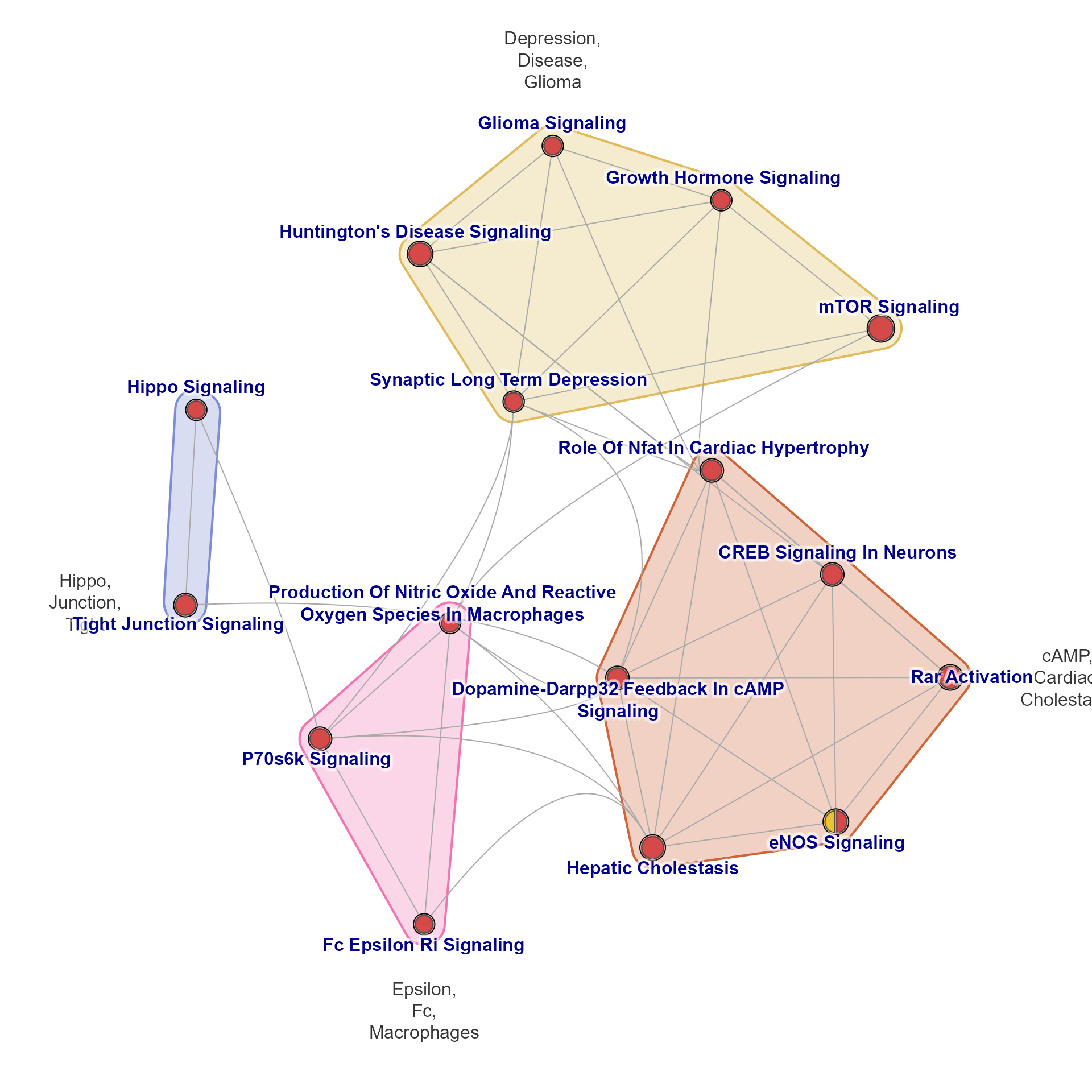
emap <- mem2emap(Memtest, min_count=2,)
jam_igraph(emap)
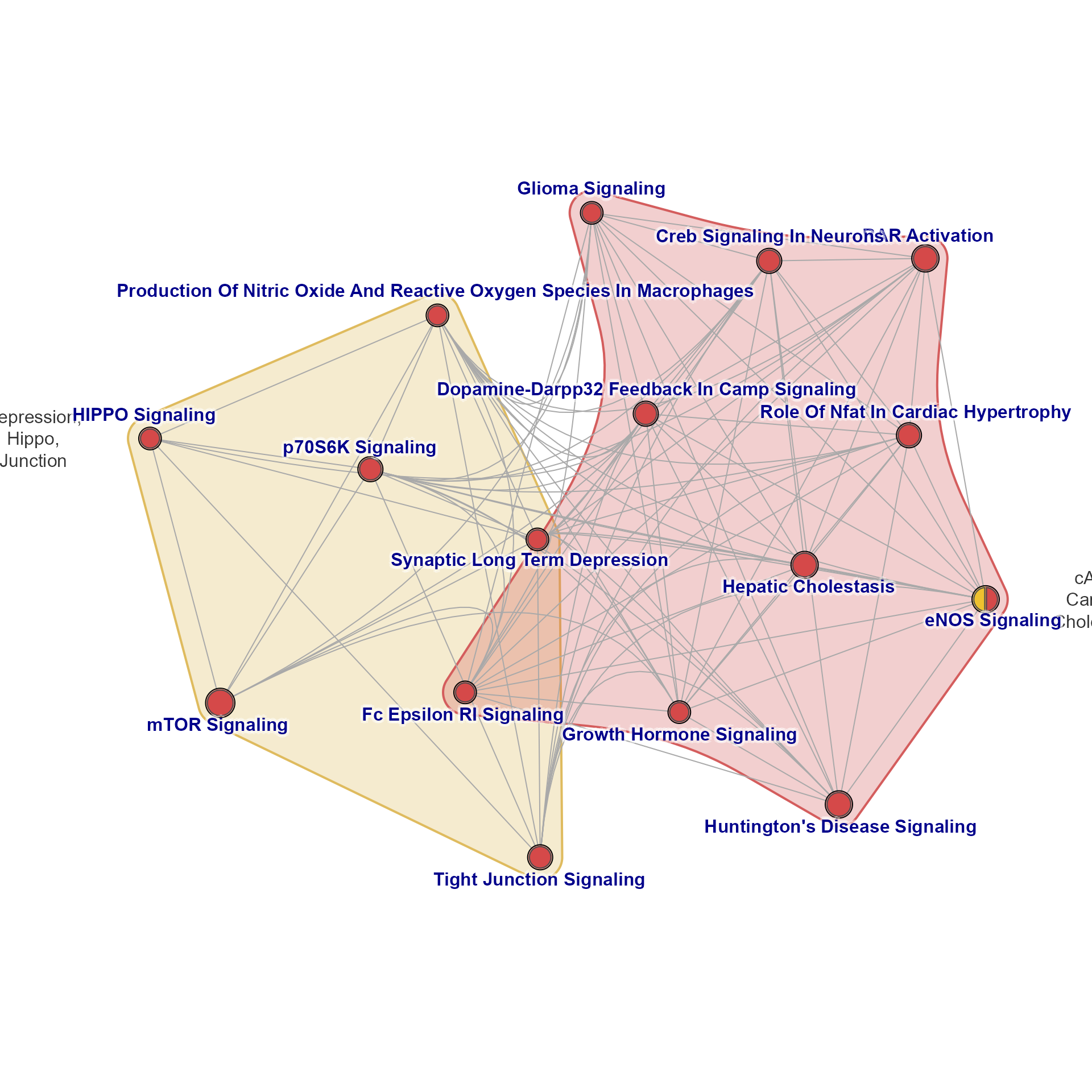 emap2 <- relayout_nodegroups(emap, final_repulse=4, y_bias=10, label_min_dist=2.5)
jam_igraph(emap2)
emap2 <- relayout_nodegroups(emap, final_repulse=4, y_bias=10, label_min_dist=2.5)
jam_igraph(emap2)
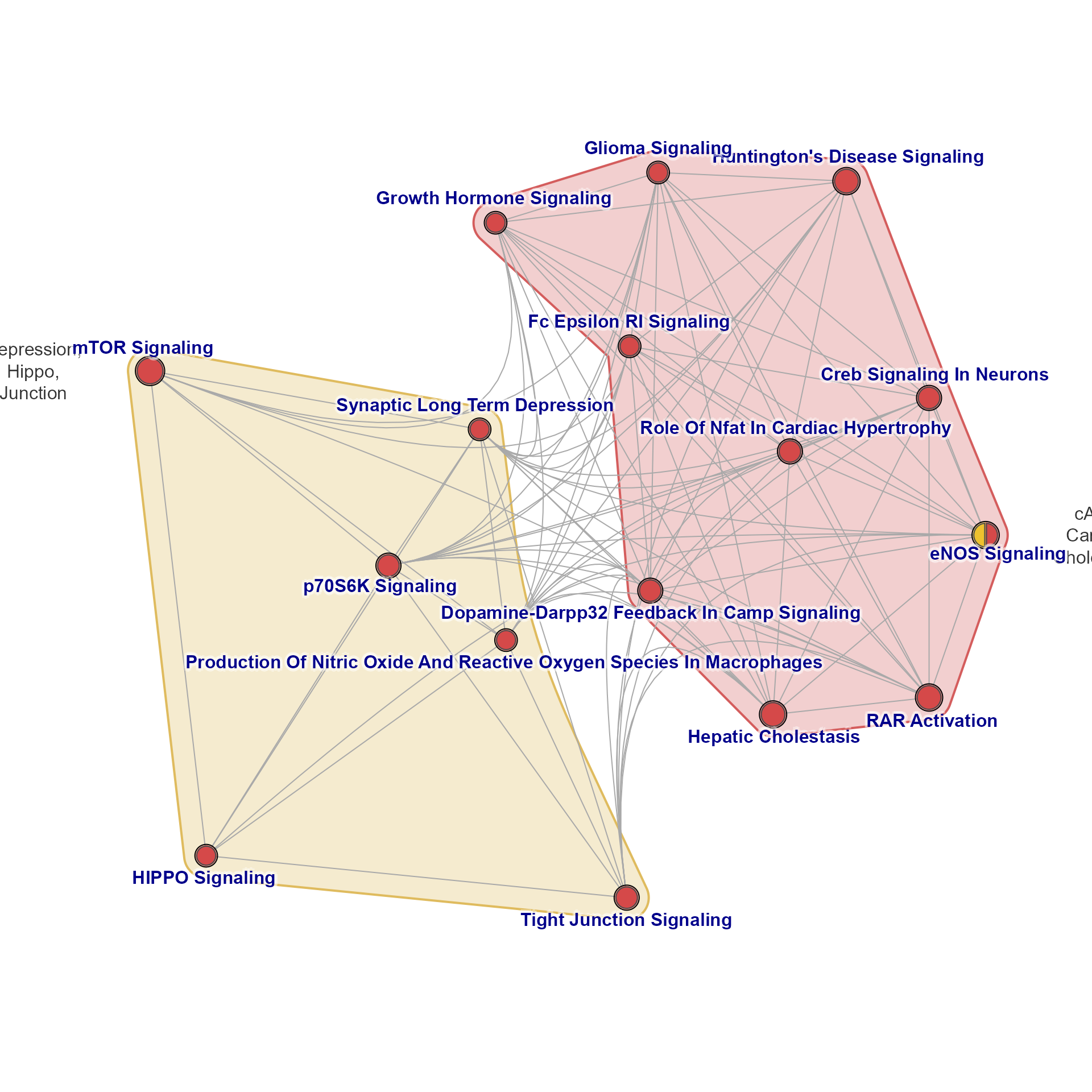 emapB <- mem2emap(fixSetLabels(Memtest), min_count=2, overlap=0.35)
jam_igraph(emapB)
emapB <- mem2emap(fixSetLabels(Memtest), min_count=2, overlap=0.35)
jam_igraph(emapB)