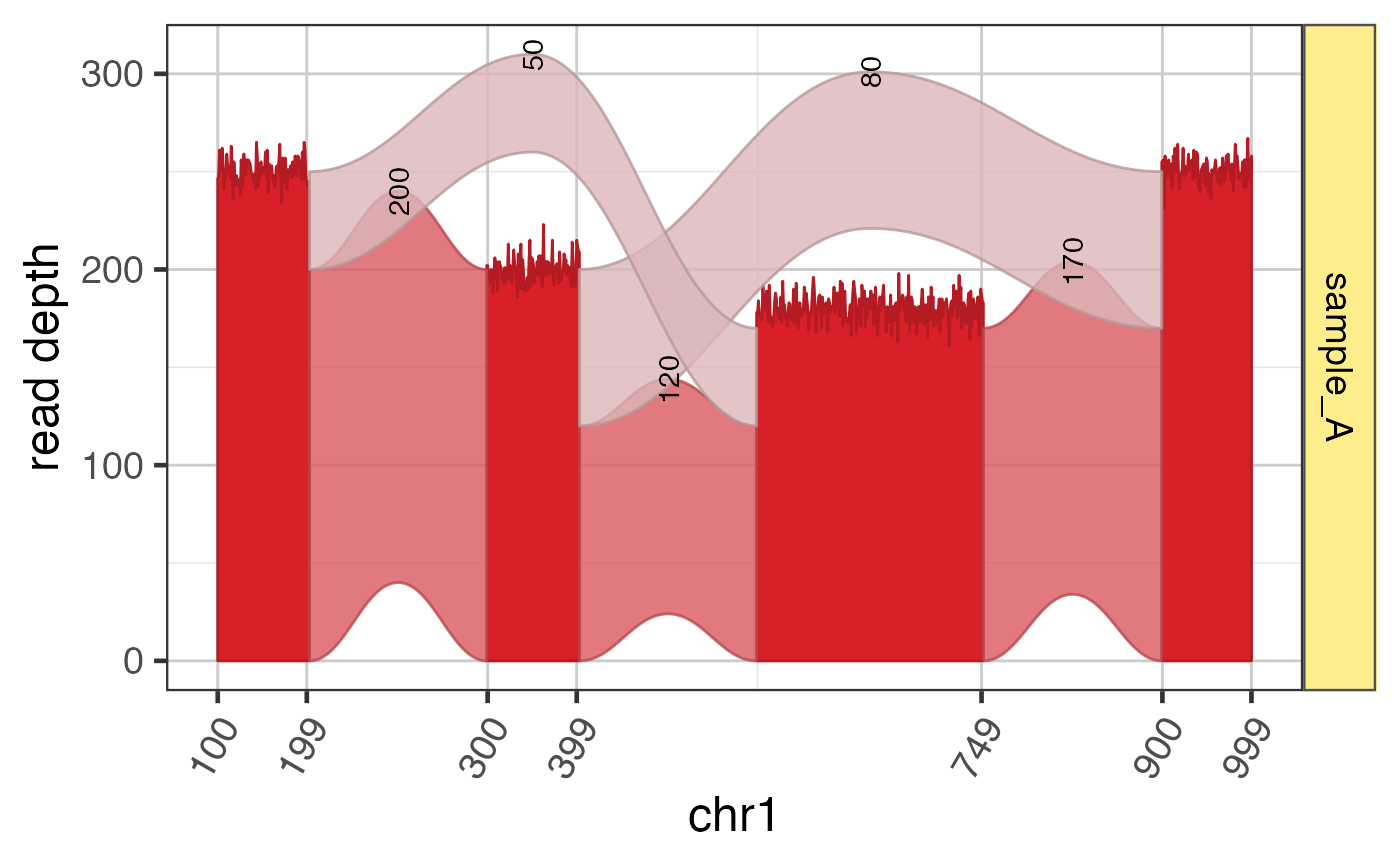
Jam Sashimi plot
plotSashimi( sashimi, show = c("coverage", "junction", "junctionLabels"), coord_method = c("scale", "coord", "none"), exonsGrl = NULL, junc_color = alpha2col("goldenrod2", 0.3), junc_fill = alpha2col("goldenrod2", 0.9), junc_alpha = 0.8, junc_accuracy = 1, fill_scheme = c("sample_id", "exon"), color_sub = NULL, ylabel = "read depth", xlabel = NULL, xlabel_ref = TRUE, use_jam_themes = TRUE, apply_facet = TRUE, facet_scales = "free_y", ref2c = NULL, label_coords = NULL, do_highlight = FALSE, verbose = FALSE, ... )
Arguments
| sashimi | Sashimi data prepared by |
|---|---|
| show | character vector of Sashimi plot features to include:
|
| coord_method | character value indicating the type of
coordinate scaling to use:
|
| exonsGrl | GRangesList object with one or more gene or transcript exon models, where exons are disjoint (not overlapping.) |
| junc_color, junc_fill | character string with valid R color,
used for junction outline, and fill, for the junction arc
polygon. Alpha transparency is recommended for |
| junc_alpha | numeric value between 0 and 1, to define the alpha transparency used for junction colors, where 0 is fully transparent, and 1 is completely non-transparent. |
| fill_scheme | character string for how the exon coverages
will be color-filled: |
| color_sub | optional character vector of R compatible colors
or hex strings, whose names are used to color or fill features
in the ggplot object. For example, if |
| ylabel | character string used as the y-axis label, by default
|
| xlabel | character string used to define the x-axis name,
which takes priority over argument |
| xlabel_ref | logical indicating whether the x-axis name should be determined by the reference (chromosome). |
| use_jam_themes | logical indicating whether to apply
|
| apply_facet | logical indicating whether to apply
|
| facet_scales | character value used as |
| ref2c | optional output from |
| label_coords | numeric vector length 2, optional range of
genomic coordinates to restrict labels, so labels are not
arranged by |
| verbose | logical indicating whether to print verbose output. |
| ... | additional arguments are sent to |
Details
This function uses Sashimi data prepared by prepareSashimi()
and creates a ggplot graphical object ready for visualization.
As a result, this function provides several arguments to
customize the visualization.
See also
Other jam plot functions:
bgaPlotly3d(),
factor2label(),
gene2gg(),
grl2df(),
jitter_norm(),
prepareSashimi(),
stackJunctions()
Other jam ggplot2 functions:
gene2gg(),
geom_diagonal_wide_arc(),
splicejam-extensions,
to_basic.GeomShape()
Other splicejam core functions:
exoncov2polygon(),
gene2gg(),
grl2df(),
make_ref2compressed(),
prepareSashimi()
Examples
suppressPackageStartupMessages(library(GenomicRanges)); data(test_exon_gr); data(test_junc_gr); data(test_cov_gr); filesDF <- data.frame(url="sample_A", type="coverage_gr", sample_id="sample_A"); sh1 <- prepareSashimi(GRangesList(TestGene1=test_exon_gr), filesDF=filesDF, gene="TestGene1", covGR=test_cov_gr, juncGR=test_junc_gr);#> Warning: invalid factor level, NA generated#> Warning: invalid factor level, NA generatedplotSashimi(sh1);