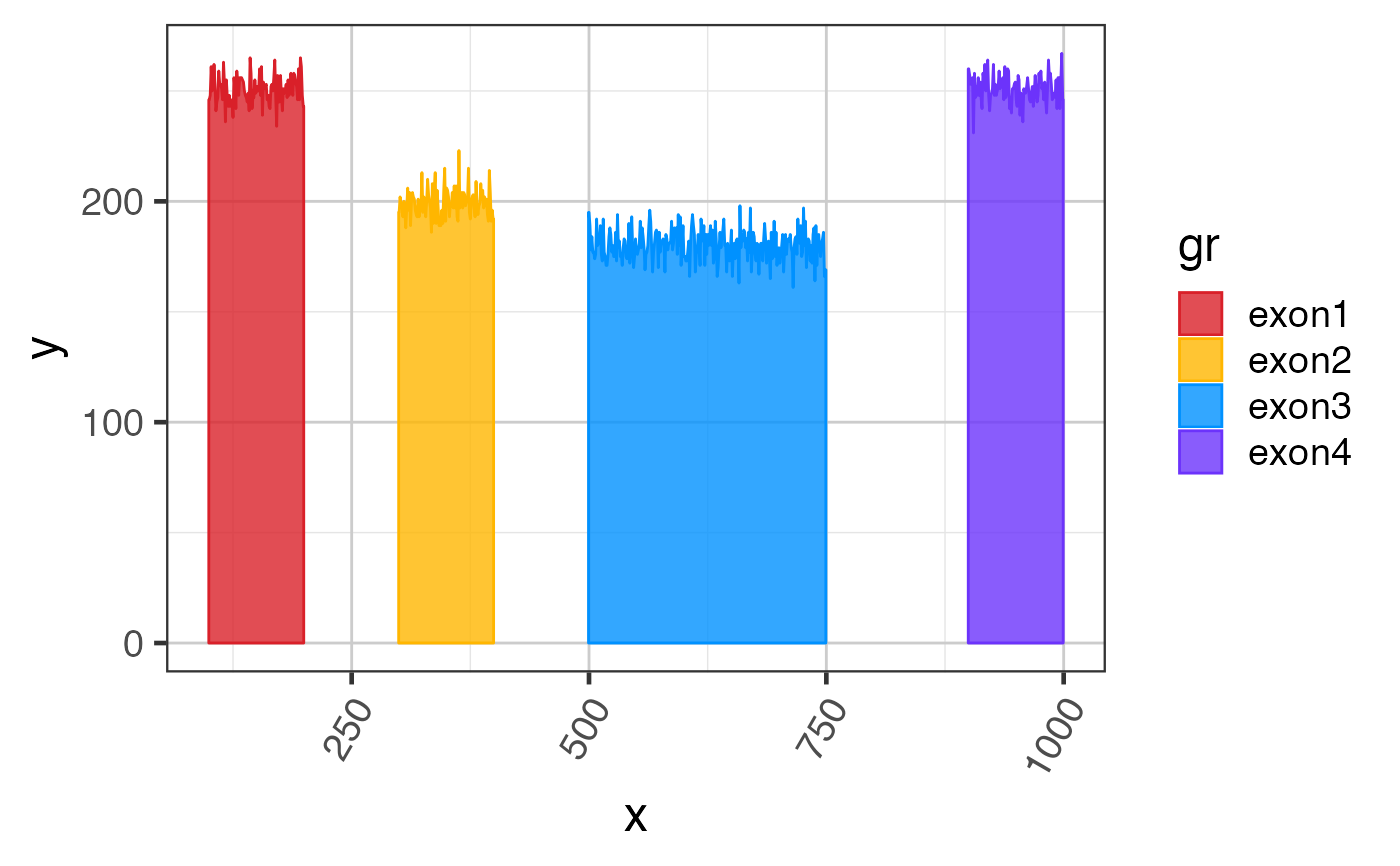
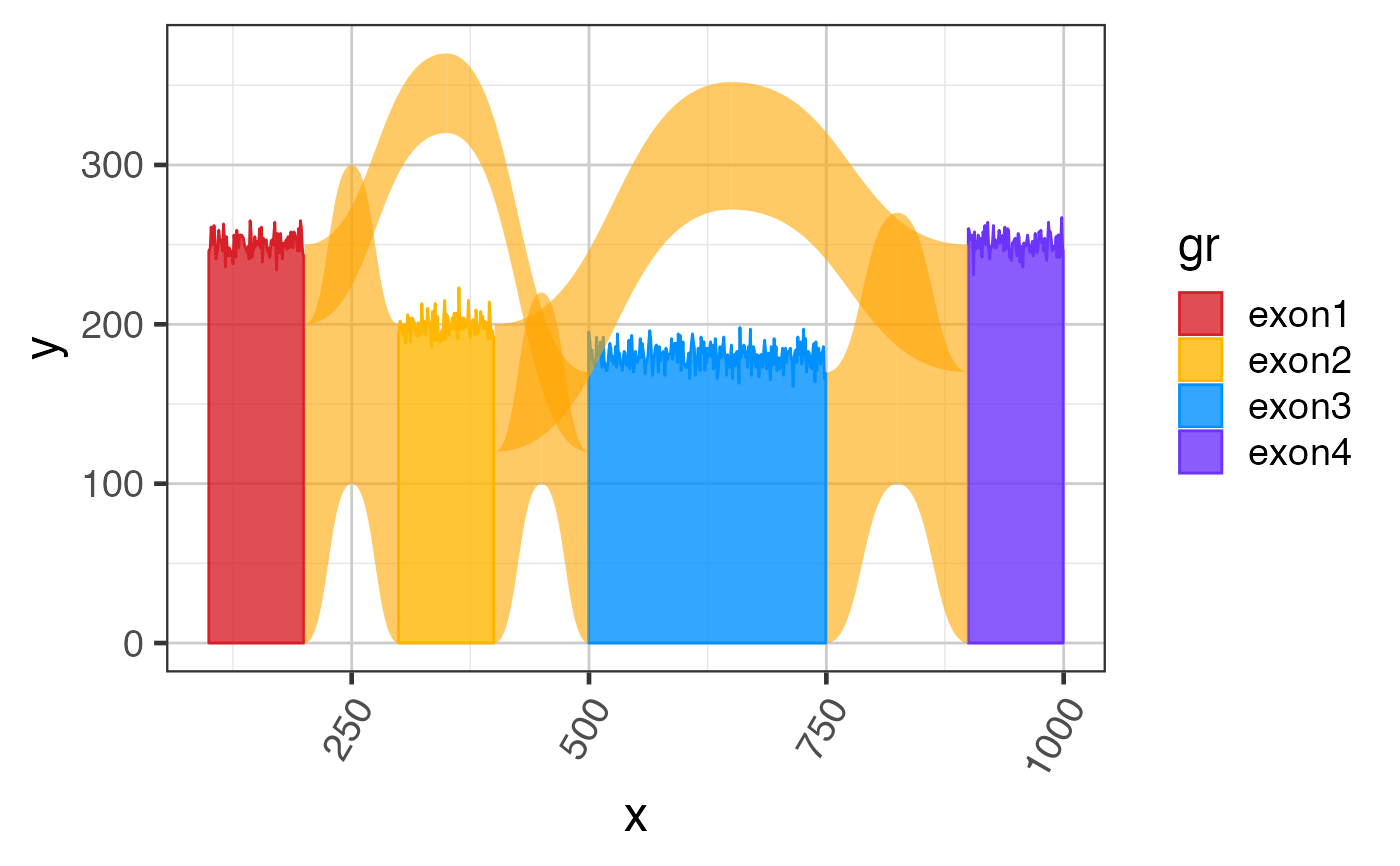
Sample exon coverage data GRanges
test_cov_gr
Format
An object of class GRanges of length 4.
Details
This dataset represents exon GRanges with an additional column with NumericList values representing RNA-seq read coverage across these exons.
See also
Other splicejam data:
test_cov_wide_gr,
test_exon_gr,
test_exon_wide_gr,
test_junc_gr,
test_junc_wide_gr
Examples
suppressPackageStartupMessages(library(GenomicRanges)); suppressPackageStartupMessages(library(ggplot2)); data(test_exon_gr); # The steps below demonstrate how to create coverage data manually test_cov_gr <- test_exon_gr; set.seed(123); cov_values <- c(250, 200, 180, 250); values(test_cov_gr)[,"sample_A"] <- NumericList( lapply(seq_along(test_exon_gr), function(i){ cov_values[i] + round(rnorm(width(test_exon_gr)[i])*7) })); test_cov_gr;#> GRanges object with 4 ranges and 2 metadata columns: #> seqnames ranges strand | gene_name sample_A #> <Rle> <IRanges> <Rle> | <character> <NumericList> #> exon1 chr1 100-199 + | TestGene1 246,248,261,... #> exon2 chr1 300-399 + | TestGene1 195,202,198,... #> exon3 chr1 500-749 + | TestGene1 195,189,178,... #> exon4 chr1 900-999 + | TestGene1 260,257,253,... #> ------- #> seqinfo: 1 sequence from an unspecified genome; no seqlengths# You can plot coverage using exoncov2polygon() exondf <- exoncov2polygon(test_cov_gr, covNames="sample_A"); gg3 <- ggplot(exondf, aes(x=x, y=y, group=gr, fill=gr, color=gr)) + ggforce::geom_shape(alpha=0.8) + colorjam::theme_jam() + colorjam::scale_fill_jam() + colorjam::scale_color_jam(); print(gg3);# you can add junctions to exons in one plot junc_df <- grl2df(test_junc_gr, shape="junction") gg3 + ggforce::geom_diagonal_wide(data=junc_df, inherit.aes=FALSE, show.legend=FALSE, fill="orange", aes(x=x, y=y, group=id), alpha=0.6)