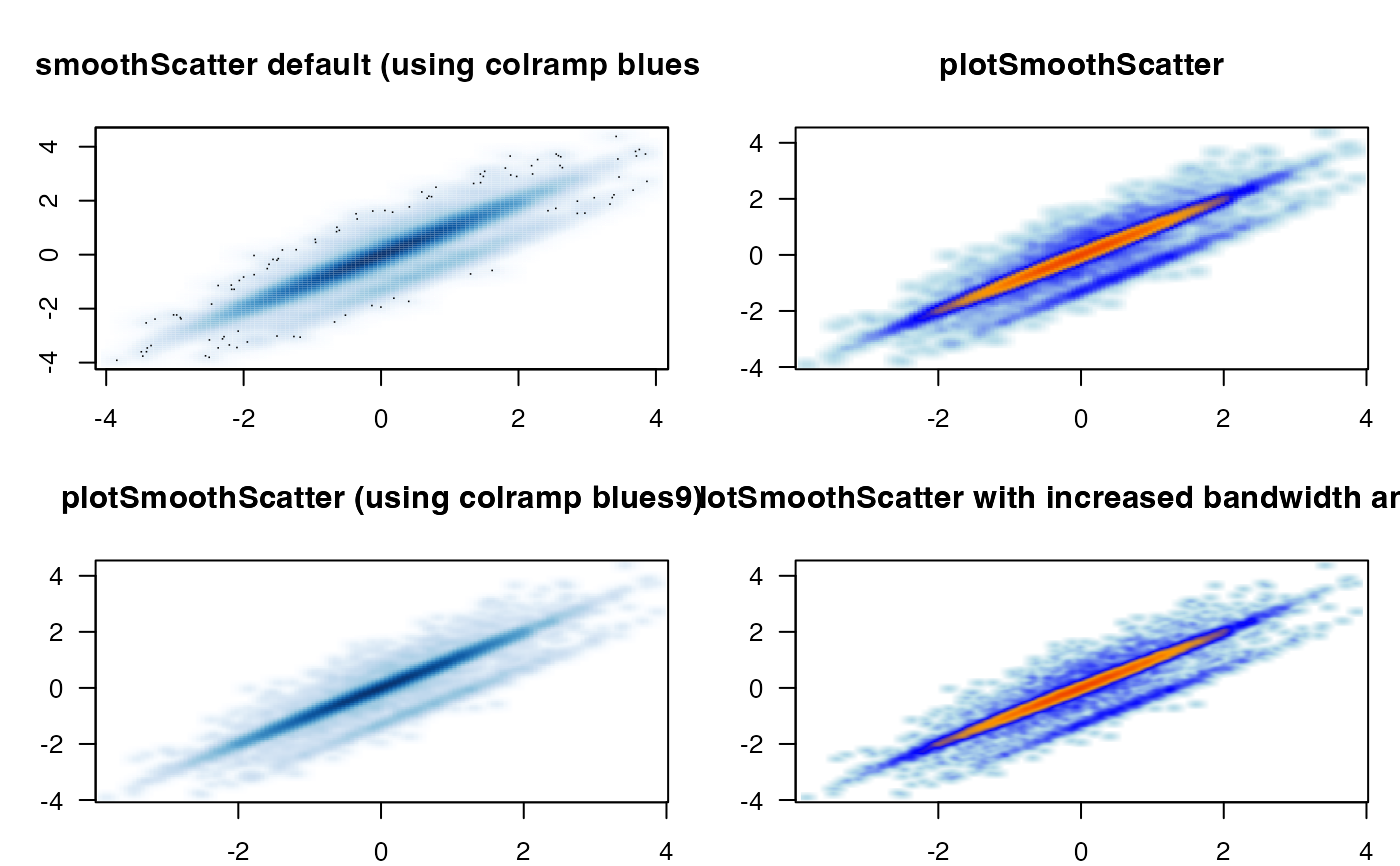
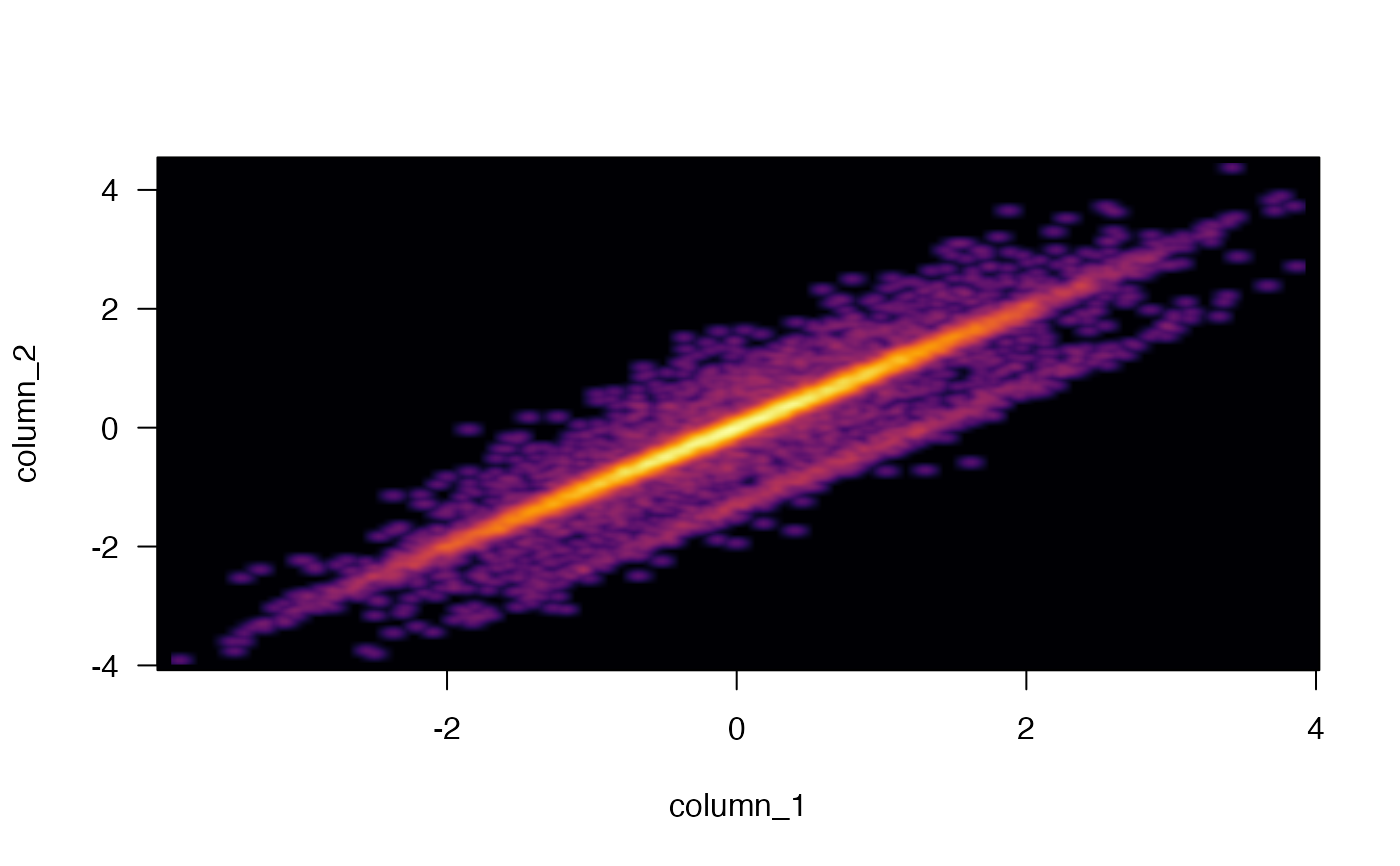
Produce scatter plot using point density instead of displaying individual data points.
Usage
plotSmoothScatter(
x,
y = NULL,
bwpi = 50,
binpi = 50,
bandwidthN = NULL,
nbin = NULL,
expand = c(0.04, 0.04),
transFactor = 0.25,
transformation = function(x) x^transFactor,
xlim = NULL,
ylim = NULL,
xlab = NULL,
ylab = NULL,
nrpoints = 0,
colramp = c("white", "lightblue", "blue", "orange", "orangered2"),
col = "black",
doTest = FALSE,
fillBackground = TRUE,
naAction = c("remove", "floor0", "floor1"),
xaxt = "s",
yaxt = "s",
add = FALSE,
asp = NULL,
applyRangeCeiling = TRUE,
useRaster = TRUE,
verbose = FALSE,
...
)Arguments
- x
numeric vector, or data matrix with two or more columns.
- y
numeric vector, or if data is supplied via x as a matrix, y is NULL.
- bwpi
numericvalue indicating the bandwidth "per inch" to scale the bandwidth based upon visual space available. This argument is used to definebandwidthN, howeverbwpiis only used whenbandwidthN=NULL. The bandwidth is used to define the 2-dimensional point density.- binpi
numericvalue indicating the number of bins "per inch", to scale based upon visual space available. This argument is used to definenbin, howeverbinpiis only used whennbin=NULL.- bandwidthN
integernumber of bandwidth steps to use across the visible plot window. Note that this bandwidth differs from defaultgraphics::smoothScatter()in that it uses the visible plot window instead of the data range, so if the plot window is not sufficiently similar to the data range, the resulting smoothed density will not be visibly distorted. This parameter also permits display of higher (or lower) level of detail.- nbin
integernumber of bins to use when converting the kernel density result (which uses bandwidthN above) into a usable image. This setting is effectively the resolution of rendering the bandwidth density in terms of visible pixels. For examplenbin=256will create 256 visible pixels wide and tall in each plot panel; andnbin=32will create 32 visible pixels, with lower detail which may be suitable for multi-panel plots. To use a variable number of bins, trybinpi.- expand
numericvalue indicating the fraction of the x-axis and y-axis ranges to add to create an expanded range, used whenadd=FALSE. The defaultexpand=c(0.04, 0.04)mimics the R base plot default which adds 4 percent total, therefore 2 percent to each side of the visible range.- transFactor
numericvalue used by the defaulttransformationfunction, which effectively scales the density of points to a reasonable visible distribution. This argument is a convenience method to avoid having to type out the fulltransformationfunction.- transformation
functionwhich converts point density to a number, typically related to square root or cube root transformation. Note that the default usestransFactorbut if a custom function is supplied, it will not usetransFactorunless specified.- xlim
numericx-axis range, orNULLto use the data range.- ylim
numericy-axis range, orNULLto use the data range.- xlab, ylab
characterlabels for x- and y-axis, respectively.- nrpoints
integernumber of outlier datapoints to display, as defined bygraphics::smoothScatter(), however the default here isnrpoints=0to avoid additional clutter in the output, and because the default argumentsbwpi,binpiusually indicate all individual points.- colramp
any input recognized by
getColorRamp():charactervector with multiple colorscharacterstring length 1, with valid R color used to create a linear color gradientcharactername of a known color gradient fromRColorBrewerorviridisfunctionthat itself produces vector of colors, in the formfunction(n)wherendefines the number of colors.
- col
characterstring with R color used whennrpointsis non-zero, this color defines the color of those points.- doTest
logicalindicating whether to create a visual set of test plots to demonstrate the utility of this function.- fillBackground
logicalindicating whether to fill the background of the plot panel with the first color incolramp. The defaultfillBackground=TRUEis useful since the plot panel may be slightly wider than the range of data being displayed, and when the first color incolrampis not the same as the plot device background color. Run a test using:plotSmoothScatter(doTest=TRUE, fillBackground=FALSE, colramp="viridis")and compare with:plotSmoothScatter(doTest=TRUE, colramp="viridis")- naAction
characterstring indicating how to handle NA values, typically when x is NA and y is not NA, or vice versa. valid values:- "remove"
ignore any points where either x or y are NA
- "floor0"
change any NA values to zero 0 for either x or y
- "floor1"
change any NA values to one 1 for either x or y
The latter two options are useful when the desired plot should indicate the presence of an NA value in either x or y, while also indicating the the corresponding non-NA value in the opposing axis. The driving use was plotting gene fold changes from two experiments, where the two experiments may not have measured the same genes.
- xaxt
charactervalue compatible with graphics::par(xaxt), used to control the x-axis range, similar to its use inplot()generic functions.- yaxt
charactervalue compatible with graphics::par(yaxt), used to control the y-axis range, similar to its use inplot()generic functions.- add
logicalwhether to add to an existing active R plot, or create a new plot window.- asp
numericwith optional aspect ratio, as described ingraphics::plot.window(), whereasp=1defines x- and y-axis coordinate ranges such that distances between points are rendered accurately. One data unit on the y-axis is equal in length toaspmultiplied by one data unit on the x-axis. Notes:When
add=TRUE, the valueaspis ignored, because the existing plot device is re-used.When
add=FALSEandaspis defined withnumericvalue, a new plot device is opened usingplot.window(), and thexlimandylimvalues are passed to that function. As a result thegraphics::par("usr")values are used to definexlimandylimfor the purpose of determining visible points, relevant toapplyRangeCeiling.
- applyRangeCeiling
logicalindicating how to handle points outside the visible plot range. Valid values:- TRUE
Points outside the viewing area are fixed to the plot boundaries, in order to represent that there are additional points outside the boundary. This setting is recommended when the reasonable viewing area is smaller than the actual data, for example to be consistent across plot panels, but where you want to indicate that points may be outside the range.
- FALSE
Points outside the viewing area is not displayed, with no special visual indication. This setting is useful when data may contain a large number of points at
c(0, 0)and the density overwhelms the detail in the rest of the plot. In that case settingxlim=c(1e-10, 10)andapplyRangeCeiling=FALSEwould obscure these points.
- useRaster
logicalindicating whether to produce plots using thegraphics::rasterImage()function which produces a plot raster image offline then scales this image to visible plot space. This technique has two benefits:It produces substantially faster plot output.
Output contains substantially fewer plot objects, which results in much smaller file sizes when saving in 'PDF' or 'SVG' format.
- verbose
logicalindicating whether to print verbose output.- ...
additional arguments are passed to called functions, including
getColorRamp(),nullPlot(),smoothScatterJam().
Value
list invisibly, sufficient to reproduce most of the
graphical parameters used to create the smooth scatter plot.
Details
This function intends to make several potentially customizable
features of graphics::smoothScatter() plots much easier
to customize. For example bandwidthN allows defining the number of
bandwidth steps used by the kernel density function, and importantly
bases the number of steps on the visible plot window, and not the range
of data, which can differ substantially. The nbin argument is related,
but is used to define the level of detail used in the image function,
which when plotting numerous smaller panels, can be useful to reduce
unnecessary visual details.
This function also by default produces a raster image plot
with useRaster=TRUE, which adjusts the x- and y-bandwidth to
produce visually round density even when the x- and y-ranges
are very different.
Comments:
asp=1will define an aspect ratio 1, meaning the x-axis and y-axis units will be the same physical size in the output device. When this is true, andfillBackground=TRUEthexlimandylimvalues follow logic forplot.default()andplot.window()such that each axis will include at least thexlimandylimranges, with additional range included in order to maintain the plot aspect ratio.When
asp, and any ofxlimorylim, are defined, the data will be "cropped" to respectivexlimandylimvalues as relevant, after which the plot is drawn with the appropriate plot aspect ratio. WhenapplyRangeCeiling=TRUE, points outside the fixedxlimandylimrange are fixed to the edge of the range, after which the plot is drawn with the requested plot aspect ratio. It is recommended not to definexlimandylimwhen also definingasp.When
add=TRUEthexlimandylimvalues are already defined by the plot device. It is recommended not to definexlimandylimwhenadd=TRUE.
See also
Other jam plot functions:
adjustAxisLabelMargins(),
coordPresets(),
decideMfrow(),
drawLabels(),
getPlotAspect(),
groupedAxis(),
imageByColors(),
imageDefault(),
minorLogTicksAxis(),
nullPlot(),
plotPolygonDensity(),
plotRidges(),
shadowText(),
shadowText_options(),
showColors(),
sqrtAxis(),
usrBox()